-
-
Notifications
You must be signed in to change notification settings - Fork 125
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Identifiers not displaying correctly #411
Comments
I see your point. This may be because the current version of the plugin is based on Jupyter Lab 1 which is really old, or it may be some oversight in the implementation of the plugin. The next version of the plugin will follow Jupyter Notebook 7 (and thus Jupyter Lab 4) closely now that PR #396 is merged. The style of the list of identifiers is what you want: However, I notice that there is still an issue in that everything is listed as "unknown", so I still need to do some work here. The docstring popup balloon is also now like in JupyterLab:
Thanks, this is good to know. I don't think I ever realized that you can right-click on the tab in Jupyter Lab. We may have to add a context menu to the tab.
I hope that we can make this automatic, so that is works just like you expected. This is the (old) issue #76. But if we can't make this work, we should give more information on the welcome screen as you suggested. |
|
@jitseniesen, I think this happens because we have Jedi completions disabled by default in Spyder: Could you check that? Thanks! |
Nop, we don't provide identifiers in the IPython console for now, sorry. |




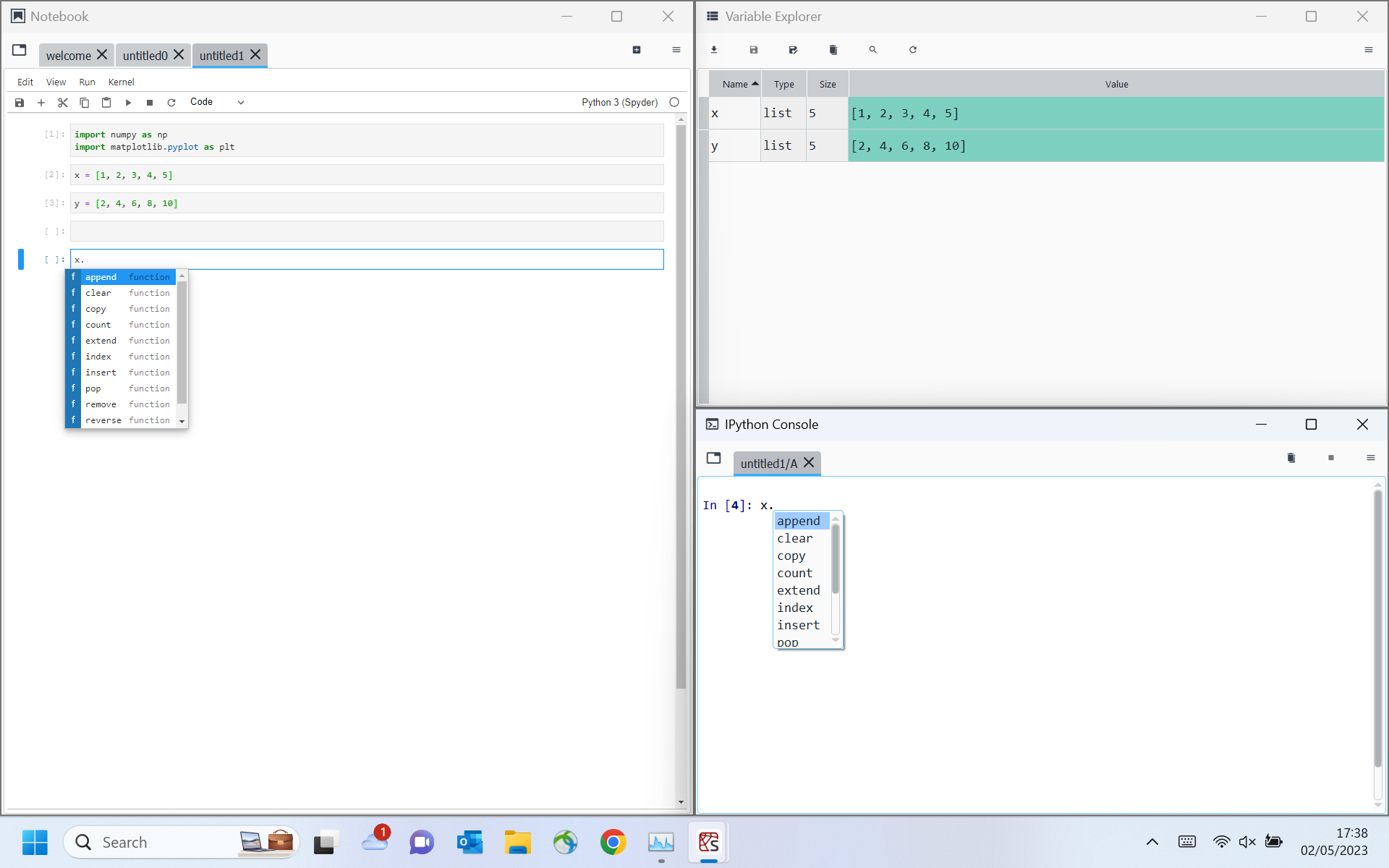
Identifiers not displaying correctly
What is the expected output? What do you see instead?
The identifiers should display as a colour-coded list either similar to JupyterLab or to those in Spyder... Not sure what style the notebook aims to follow... Currently the ↹ needs to be selected to view the list of identifiers, ⇧ and ↹ need to be pressed to display the docstring popup balloon. This behaviour is more similar to JupyterLab and the docstring itself uses the style format of JupyterLab.
Personally I prefer the style of identifiers and docstrings in JupyterLab and would rather have the ability to use this style exclusively throughout Spyder in the Script Editor, Notebook and Consoles. They are a bit cleaner and easier to read.
Some other things I noticed were that right clicking the tab gave no file context menu (similar to JupyterLab). A subset of these actions seem to be in a separate Options Menu.
Also usability wise, this was the first time I used the notebook extension itself but have used JupyterLab and Spyder quite extensively. The Welcome to Spyder-Notebook needs a bit more information:
This is not enough to figure out that:
Details about this are on the home page but not enough details are on the welcome tab to figure out this procedure.
Please provide any additional information below (stack trace, screenshots, a gif, etc)
List of Identifiers in Spyder Notebook:
List of Identifiers in JupyterLab:
List of Identifiers in Spyder:
Docstring Popup Balloon in Spyder Notebook:
Docstring Popup Balloon in JupyterLab:
Docstring Popup Balloon in Spyder:
JupyterLab Tab Menu:
Spyder Notebook Option Menu:
Welcome:
New Notebook:
Open Console:
Work with Interactive Notebook:
Update IPython Console:
Versions and main components
Dependencies
Mandatory:
atomicwrites >=1.2.0 : 1.4.0 (OK)
chardet >=2.0.0 : 4.0.0 (OK)
cloudpickle >=0.5.0 : 2.0.0 (OK)
cookiecutter >=1.6.0 : 1.7.3 (OK)
diff_match_patch >=20181111 : 20200713 (OK)
intervaltree >=3.0.2 : 3.1.0 (OK)
IPython >=7.31.1,<9.0.0,!=8.8.0,!=8.9.0,!=8.10.0 : 8.12.0 (OK)
jedi >=0.17.2,<0.19.0 : 0.18.1 (OK)
jellyfish >=0.7 : 0.9.0 (OK)
jsonschema >=3.2.0 : 4.17.3 (OK)
keyring >=17.0.0 : 23.13.1 (OK)
nbconvert >=4.0 : 6.5.4 (OK)
numpydoc >=0.6.0 : 1.5.0 (OK)
paramiko >=2.4.0 : 2.8.1 (OK)
parso >=0.7.0,<0.9.0 : 0.8.3 (OK)
pexpect >=4.4.0 : 4.8.0 (OK)
pickleshare >=0.4 : 0.7.5 (OK)
psutil >=5.3 : 5.9.0 (OK)
pygments >=2.0 : 2.11.2 (OK)
pylint >=2.5.0,<3.0 : 2.16.2 (OK)
pylint_venv >=2.1.1 : 2.3.0 (OK)
pyls_spyder >=0.4.0 : 0.4.0 (OK)
pylsp >=1.7.2,<1.8.0 : 1.7.2 (OK)
pylsp_black >=1.2.0 : 1.2.1 (OK)
qdarkstyle >=3.0.2,<3.2.0 : 3.0.2 (OK)
qstylizer >=0.2.2 : 0.2.2 (OK)
qtawesome >=1.2.1 : 1.2.2 (OK)
qtconsole >=5.4.2,<5.5.0 : 5.4.2 (OK)
qtpy >=2.1.0 : 2.2.0 (OK)
rtree >=0.9.7 : 1.0.1 (OK)
setuptools >=49.6.0 : 65.6.3 (OK)
sphinx >=0.6.6 : 5.0.2 (OK)
spyder_kernels >=2.4.3,<2.5.0 : 2.4.3 (OK)
textdistance >=4.2.0 : 4.2.1 (OK)
three_merge >=0.1.1 : 0.1.1 (OK)
watchdog >=0.10.3 : 2.1.6 (OK)
zmq >=22.1.0 : 23.2.0 (OK)
Optional:
cython >=0.21 : 0.29.33 (OK)
matplotlib >=3.0.0 : 3.7.1 (OK)
numpy >=1.7 : 1.23.5 (OK)
pandas >=1.1.1 : 2.0.0 (OK)
scipy >=0.17.0 : 1.10.1 (OK)
sympy >=0.7.3 : 1.11.1 (OK)
Spyder plugins:
spyder_notebook.notebookplugin 0.4.1 : 0.4.1 (OK)
The text was updated successfully, but these errors were encountered: